Pushkar Malakar
Dept. of Biomedical Science and Technology,
School of Biological Sciences,
RKMVERI, Belur Math, Howrah-711202
West Bengal, India
Contact: +91-6289799952
Email:
Brahmachari Vedeshachaitanya (Somnath), Ph.D.
Dept. of Biomedical Science and Technology,
School of Biological Sciences,
RKMVERI, Belur Math, Howrah-711202
West Bengal, India
Email:
Research Interests
- Signaling pathways in Cancer
- Cancer Metabolism
- Cancer Drug Resistance
- Long Noncoding RNAs
- Micro RNAs in Cancer
- Polyploidy in Cancer
Ongoing projects
- Functional Roles of Long Noncoding RNAs in Physiological and Pathological Processes
- Molecular and Cellular Characterization of Polyploidy in Cancer
- Role of Glutamine Metabolism in Cancer Progression
- Mechanistic Basis of Chemoresistance in Cancer, specially in Multiple myeloma
Ongoing funding
- Ramanujan Fellowship:
Role of long noncoding RNAs in physiological and pathological conditions (2022-2027) - ANRF-ARG
Understanding the Mechanisms of Chemoresistance to Bortezomib in Multiple Myeloma and Exploring Therapeutic Strategies to Enhance Drug Sensitivity (2026-2029)
Ph.D. Students
- Didhiti Singha
- Debopriya Choudhury
- Jahir Hossain
- Soham Bhattacharyya
Publications
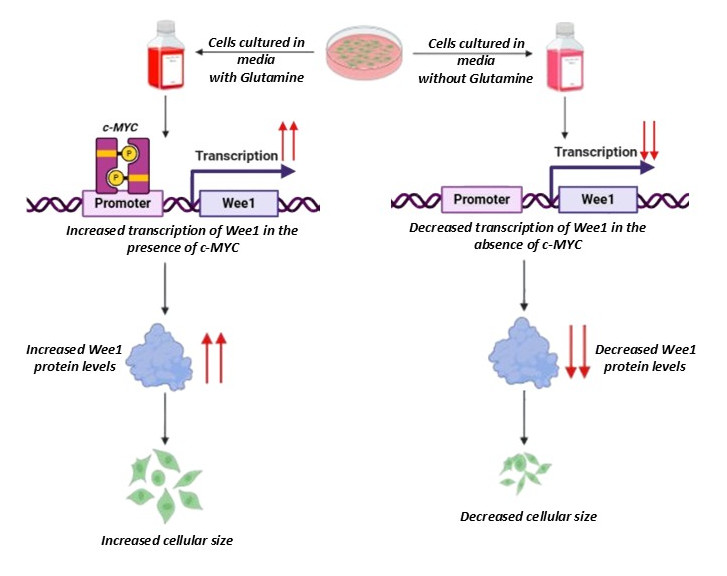
- Glutamine modulates cellular size by regulating Wee1 expression. Didhiti Singha, Meghna Mondal, Debopriya Choudhury, and Pushkar Malakar. Scientific Reports.2025. DOI is 10.1038/s41598-025-17687-7. As Corresponding Author.
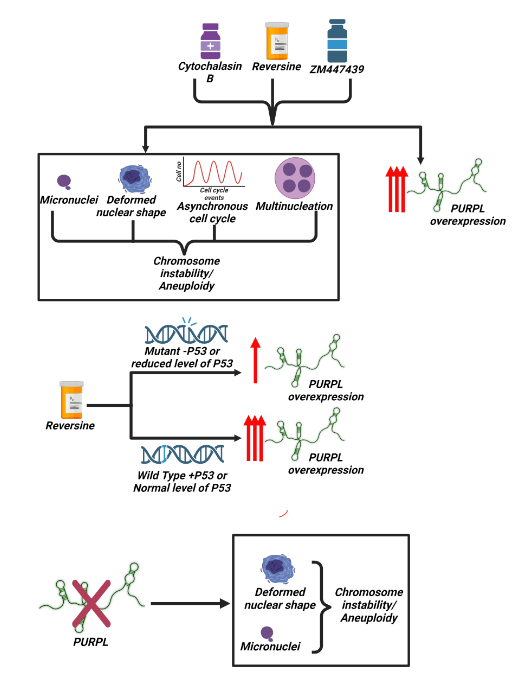
- Long noncoding RNA, PURPL is associated with aneuploidy and its magnitude of expression level is dependent on P53 status. Malakar P ##. Front Cell Dev Biol. 2024 Dec 23; 12:1410308. # #As Single and Corresponding Author.
- Unleashing innovation: 3D-printed biomaterials in bone tissue engineering for repairing femur and tibial defects in animal models – a systematic review and meta-analysis. Sagar N, Chakravarti B, Maurya SS, Nigam A, Malakar P, Kashyap R. Front Bioeng Biotechnol. 2024 Sep 23; 12:1385365.
- Microplastics in the soil-water-food nexus: Inclusive insight into global research findings. Garai S, Bhattacharjee C, Sarkar S, Moulick D, Dey S, Jana S, Dhar A, Roy A, Mondal K, Mondal M, Mukherjee S, Ghosh S, Singh P, Ramteke P, Manna D, Hazra S, Malakar P, Banerjee H, Brahmachari K, Hossain A. Sci Total Environ. 2024 Oct 10; 946:173891.
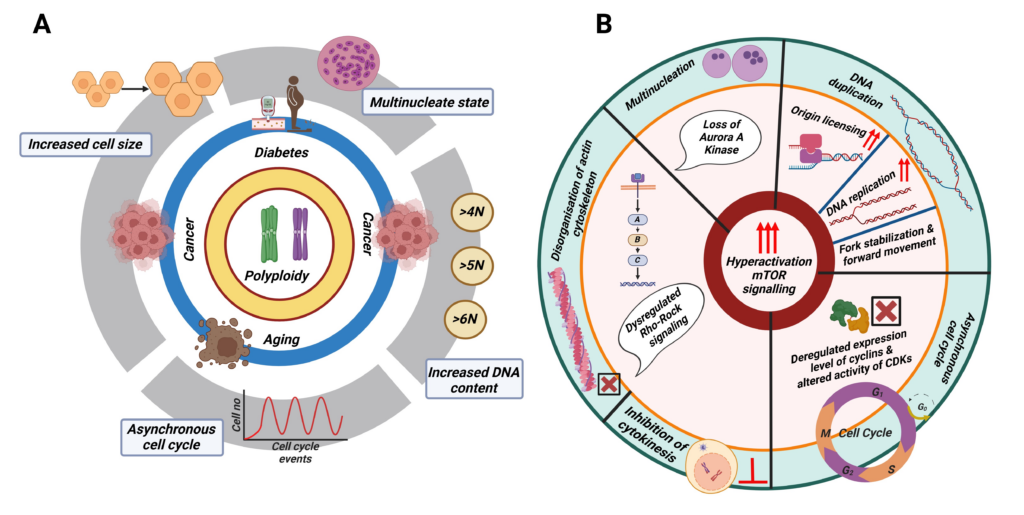
- Characterization of polyploidy in cancer: Current status and future perspectives. Ghosh S, Choudhury D, Ghosh D, Mondal M, Singha D, Malakar P *. Int J Biol Macromol. 2024 May;268(Pt 1):131706. * As Corresponding Author.
- Polyploidy and mTOR signaling: a possible molecular link. Choudhury D, Ghosh D, Mondal M, Singha D, Pothuraju R, Malakar P *. Cell Commun Signal. 2024 Mar 27;22(1):196. * As Corresponding Author.
- Gum acacia dietary fiber: Significance in immunomodulation, inflammatory diseases, and cancer. Barkeer S, Pothuraju R, Malakar P, Pimentel TC, Siddiqui JA, Nair SA. Phytother Res. 2024 Mar;38(3):1509-1521.
- The nexus of long noncoding RNAs, splicing factors, alternative splicing and their modulations. Malakar P#, Shukla S, Mondal M, Kar RK, Siddiqui JA. RNA Biol. 2024 Jan;21(1):1-20. # As first and corresponding author
- The role of alternative splicing and splicing factors in diabetes: Current status and future perspectives. Singha D, Mondal M, Ghosh D, Choudhury D, Chakravarti B, Kar RK, Malakar P*. Wiley Interdiscip Rev RNA. 2024;15(1): e1831. *As Corresponding Author.
- Pan-cancer integrated bioinformatic analysis of RNA polymerase subunits reveal RNA Pol I member CD3EAP regulates cell growth by modulating autophagy. Bhandari N, Acharya D, Chatterjee A, Mandve L, Kumar P, Pratap S, Malakar P, Shukla SK. Cell Cycle. 2023 Oct 5:1-17.
- A Possible Novel Therapeutic Targets of Selinexor in Acute Lymphoblastic Leukemia: A Comprehensive Review. Malakar P #, Sagar N, Chakravarti B, Mondal M, Singha D, Kar R. J Clin Transl Res. 2023;9(6):423-432. # As First and Corresponding Author
- Giant cells: multiple cells unite to survive. Hazra S, Kalyan Dinda S, Kumar Mondal N, Hossain SR, Datta P, Yasmin Mondal A, Malakar P, Manna D. Front Cell Infect Microbiol. 2023 Sep 5; 13:1220589.
- Glutamine regulates the cellular proliferation and cell cycle progression by modulating the mTOR mediated protein levels of β-TrCP. Malakar P#, Singha D, Choudhury D, Shukla S. Cell Cycle. 2023 Sep 28:1-14. # As First and Corresponding Author
- Molecular signaling network and therapeutic developments in breast cancer brain metastasis. Benjamin M, Malakar P, Sinha RA, Nasser MW, Batra SK, Siddiqui JA, Chakravarti B. Adv Cancer Biol Metastasis. 2023 Jul; 7:100079
- Somnath Ghosal and Subrata Banerjee. In silico bioinformatics analysis for identification of differentially expressed genes and therapeutic drug molecules in Glucocorticoid-resistant Multiple myeloma. Medical Oncology, 2022;39(5):53. https://doi.org/10.1007/s12032-022-01651-w.
- Saran Chattopadhyaya* and Somnath Ghosal*. DNA methylation: a saga of genome maintenance in hematological perspective. Human Cell, 2022;35(2):448-461. (*Equal Contribution). https://doi.org/10.1007/s13577-022-00674-9.
- Somnath Ghosal and Subrata Banerjee. Investigating the potential molecular players and therapeutic drug molecules in Carfilzomib resistant Multiple myeloma by comprehensive bioinformatics analysis. Leukemia & Lymphoma, 2022;1-12. https://doi.org/10.1080/10428194.2022.2087064.
- Somnath Ghosal and Subrata Banerjee. Identification of Differentially Expressed Genes and Therapeutic Drug Molecules in Glucocorticoid Resistant Multiple Myeloma by In Silico Bioinformatics Analysis. Indian Journal of Hematology and Blood Transfusion, 2021;37(Suppl 1):1-172. https://doi.org/10.1007/s12288-021-01510-0.
- Somnath Ghosal, Saran Chattopadhyaya and Subrata Banerjee. In silico and in vitro analysis of differentially expressed micro RNAs, circular RNAs and p53 in Bortezomib resistant Multiple myeloma. Biomedical Research and Therapy, 2022; 9(7), 5179-5190. https://doi.org/10.15419/bmrat.v9i7.754.
- Somnath Ghosal and Subrata Banerjee. Identification of potential molecular players and therapeutic drug molecules in Melphalan resistant Multiple myeloma by integrated bioinformatics analysis. Research Journal of Biotechnology, 2022; 17(12), 6-15. https://doi.org/10.25303/1712rjbt06015.
- Emerging frontiers of antibiotics use and their impacts on the human gut microbiome. Kumari R, Yadav Y, Misra R, Das U, Das Adhikari U, Malakar P, Dubey GP. Microbiol Res. 2022 Oct; 263:127127. (7 Citations).
- Long Noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR mediated TCF7L2 translation. Malakar P, Stein I, Saragovi A, Winkler R, Stern-Ginossar N, Berger M, Pikarsky E, Karni R. Cancer Res. 2019 May 15;79(10):2480-2493. (141 Citations).
- Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, Karni R. Cancer Res. 2017 Mar 1;77(5):1155-1167. (265 Citations). Most cited paper from the postdoc lab in the span of 16 years.
- Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, Jensen T, Prasanth SG, Karni R, Ray PS, Prasanth KV. Oncotarget. 2016 Jun 28;7(26):40418-40436. (149 Citations).
- Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival. Malakar P, Chartarifsky L, Hija A, Leibowitz G, Glaser B, Dor Y, Karni R. Sci Rep. 2016 Aug 16; 6: 31222. (73 Citations).
- Effect on β-galactosidase synthesis and burden on growth of osmotic stress in Escherichia coli. Malakar P #, Singh VK, Karmakar R, Venkatesh KV. Springerplus. 2014 Dec 17; 3:748. (14 Citations). # As Corresponding Author.
- Characterization of cost with respect to nutritional upshift in the media composition along with sublethal doses of transcriptional and translational inhibitor. Malakar P##. Arch Microbiol. 2014 Apr; 196(4):289-94. (4 Citations). # #As Single and Corresponding Author.
- GAL regulon of Saccharomyces cerevisiae performs optimally to maximize growth on galactose. Malakar P, Venkatesh KV. FEMS Yeast Res. 2014 Mar; 14(2):346-56. (19 Citations).
- Characterization of burden on growth due to the nutritional state of media and pre-induced gene expression. Malakar P, Venkatesh KV. Arch Microbiol. 2013 Apr;195(4):291-5. (6 Citations).
- Effect of substrate and IPTG concentrations on the burden to growth of Escherichia coli on glycerol due to the expression of Lac proteins. Malakar P, Venkatesh KV. Appl Microbiol Biotechnol. 2012 Mar; 93(6):2543-9. (115 Citations). Second most cited paper from the lab in a span of 25 years.
- Stability analysis of the GAL regulatory network in Saccharomyces cerevisiae and Kluyveromyces lactis. Kulkarni VV, Kareenhalli V, Malakar P, Pao LY, Safonov MG, Viswanathan GA. BMC Bioinformatics. 2010 Jan 18; 11 Suppl 1: S43. (18 Citations).
- Comparative modeling and genomics for galactokinase (Gal1p) enzyme. Sharma A, Malakar P#. Bioinformation. 2011 Feb 15; 5(10):422 -429. (1 Citations). # As Corresponding Author.
- Structure modeling and comparative genomics for epimerase enzyme (Gal10p). Sharma A, Malakar P#. Bioinformation. 2010 Nov 27; 5(6):266-70. (2 Citations). # As Corresponding Author
- Prospective of the application of synthetic biology to engineered genetic constructs. Pushkar Malakar##. Research and Review: A journal of microbiology and Virology. Volume 2, Issue 2, August 2012, Pages1-13. # #As Single and Corresponding Author.
- Prospective of the application of synthetic biology to engineered genetic constructs. Pushkar Malakar##. Research and Review: A journal of microbiology and Virology. Volume 2, Issue 2, August 2012, Pages1-13. # #As Single and Corresponding Author.
- Structure Modelling of Transferase (Gal7p) Protein from S.cerevisiae and K.lactis for regulation of Galactose pathway. Pushkar Malakar# and Ashwani Sharma. Journal of Advanced Bioinformatics Application and Research. Vol 1, Issue 1, June 2010, pp 41-49. # As Corresponding Author
Awards and recognitions of research group
International Awards:
- Post –Doctoral Fellowship from NIH, USA 2019.
- Travel Award from RNA society for attending RNA2017.
- Travel Award from Hebrew University for attending RNA 2017.
- Second Prize of the competition for Outstanding Papers 2016-2017, Department of Biochemistry and Molecular Biology, IMRIC, Hebrew University.
- Work selected for Oral presentation at Regulatory & Non-Coding RNAs meeting, CSHL Meetings, CSHL, USA, August23-27, 2016.
- 2014 ISCR Best Abstract Award and selected for oral presentation: Sixth Annual Meeting of the Israeli Society for Cancer Research-2014.
- Post –Doctoral Fellowship from Israeli Government (PBC) 2013.
- Silver Medal in iGEM-2009 (International Synthetic Biology Competition).
- Marie Curie Fellowship (for training on system and synthetic biology) from September-November 2008.
National Awards:
- Best poster award at Frontiers in Modern Biology (FIMB-2026) IISER Kolkata
- One works got selected for Oral presentation at 8th Regional Science and Technology Congress, Organized by Department of Science & Technology and Biotechnology, Govt. of West Bengal 2026
- Two best poster awards at International Conference on Cutting -Edge Cancer Research & Innovations for Improving Patient Outcomes (NCCS-Pune) 2025
- CSIR SRF award 2025
- Two works got selected for Oral presentation at 7th Regional Science and Technology Congress, Organized by Department of Science & Technology and Biotechnology, Govt. of West Bengal 2025
- First Prize in Oral Presentation at International seminar on the present status of cancer and other emerging diseases, organized by Ramakrishna Mission Vivekananda Centenary College, Rahara, 2024.
- Third Prize in Poster Presentation at International seminar on the present status of cancer and other emerging diseases, organized by Ramakrishna Mission Vivekananda Centenary College, Rahara, 2024.
- Ramanujan Fellowship from DST-SERB 2022.
- CSIR Travel Grant for Attending ASM (American Society for Microbiology) 2012.
- DST Travel Grant for Attending ASM (American Society for Microbiology) 2012.
- DBT- CTEP Travel Grant for Attending ASM (American Society for Microbiology) 2012.
- Award for Oral Presentation at Biodesign India, University of Kerala
- CSIR-NET JRF (Fellowship for doing Ph.D.) December 2007.
- DBT-JRF Fellowship (Fellowship for doing Ph.D.) May 2006 (Rank 39th).
- GATE Fellowship (Fellowship for doing M.Tech /Ph.D.) February 2006 (Rank 31st).
- CSIR-NET JRF (Fellowship for doing Ph.D.) December 2005.
- M.Sc. (Biotechnology) Fellowship 2004.
Lab Alumni
- Subhabrata Dey (B.Sc. Intern-Currently Masters student at The University of Salford, UK)
- Meghna Mondal (M.Sc. Project)
- Aroni Chatterjee (Project Staff-Currently Assistant Professor at Brainware University, Kolkata)
- Srijonee Ghosh (M.Sc. Project-Currently working in Biotech Industry)
- Madhumita Biswas (M.Sc. Project)
- Tanmoy Das (M.Sc. Project)
- Dhruba Ghosh (Project Staff)
- Newton Mondol (B.Sc. Intern-Currently doing M.Sc. Zoology at BHU, Varanasi)
- Debarpon Mondal (B.Sc-Intern-Currently an Integrated Ph.D. student at JNCASR, Bangalore)
- Subhadip Bhaskar (B.Sc. Intern)
- Srijita Ganguly (B.Sc. Intern)
- Rivu Chakravarty (M.Sc. Project)
- Smarta Das (M.Sc. Project)